Peptide Antigen Design

We offer a complementary peptide design tool to generate custom antibodies with better performance.
The LifeTein Protein Analysis Tool is a free bioinformatics protein sequence analysis software program. It incorporates exclusive and powerful bioinformatics algorithms to design and analyze peptide antigens to ensure that each project results in a custom highly specific antibody with a high-titer.
The Protein Analysis Tool is a web-based software program that calculates the chemical formula of a peptide, as well as its molecular weight, net charge at neutral pH, hydroelectric point, hydrophilicity and hydrophobicity, and extinction coefficient. The program can accurately predict important structural characteristics of the input protein and peptide sequences such as hydrophobic or hydrophilic regions, secondary structure, transmembrane regions, flexible regions, and the surface probability.
Applications
Peptides are complex biological molecules with unique physical and chemical properties, which result directly from their amino acid composition. Peptides can be designed either de novo, or can be based on the sequences of native proteins, depending on the final application. To maximize the probability that antibodies directed against synthesized peptides will recognize native proteins in target assays, a suitable peptide sequence must be selected for antigen design. Below, we outline a number of important principles that should be considered during the peptide design process. The technical support department at LifeTein can also assist you with peptide design.
Generally, a good peptide antigen is derived from a sequence located on the surface of the native protein. An optimal sequence has a flexible structure (usually a loop) with a unique sequence, is easy to synthesize, and possesses no sited that are modified post-translationally.
Homology Considerations:
When designing a peptide antigen, two basic strategies should be considered:
- Select a unique sequence to help ensure specificity against the target protein.
- Select a homologous peptide sequence to allow a single antibody to recognize multiple target proteins.
Epitope Selection:
Generally, most optimal antigenic epitopes are flexible, hydrophilic, located on the surface of the protein. This is because hydrophilic protein regions are commonly located on the surface of proteins, whereas hydrophobic regions tend to be inside the protein in most natural environments. In addition, antibodies can bind only to epitopes that are located on a protein surface, and generally bind with higher affinity when the epitopes are flexible and readily move into most accessible positions.
Continuous Epitopes, Discontinuous Epitopes:
X-ray crystallography can be used to determine the structure of proteins, which helps scientists visualize macromolecular protein complexes, predict the effect of specific amino acid mutations, and also design targeted therapeutic agents. However, proteins are generally more flexible that their three-dimensional structures would suggest. Intracellular proteins fold and unfold continuously. Most antibodies target a continuous amino acid sequence, or continuous epitopes. Antibodies bind to these regions with high affinity if those sequences are located toward the surface of the protein. However, antibodies can also be generated against discontinuous epitopes in instances where the epitopes are against a fold within a peptide sequence, or in a location where two distinct peptide chains meet. However, the peptide used for immunization must have a similar secondary structure to the epitope for this to be successful.

If the final research application focuses on specific protein domains, such as the N- and C-termini, or a specifically altered protein state, such as phosphorylation, the peptide antigens and the resulting antibodies are relatively easy to use. However, the conformation of target proteins might prevent the antibody from accessing the target epitopes. For example, a particular sequence could become inaccessible if it is hidden inside of a folded protein. Therefore, this is an important consideration during peptide design for antibody recognition purposes.
Targeting the N-terminus or C-terminus:
To reduce the chances of designing an epitope buried within a protein, we generally recommend that antibodies are designed against the C- or N-terminal regions of proteins, which are often exposed. However, it is important to note that transmembrane domains and the C-terminal sequences of many membrane proteins are commonly too hydrophobic to be used as antigens.

Sequence Length:
Generally, we recommend that peptide sequences of 8-20 amino acids should be used for antibody preparation. If the peptide is too short, it is unlikely to be specific enough to allow the resulting antibodies to recognize the target native protein with sufficient affinity. Conversely, sequences longer than 20 amino acids might lose their specificity and induce secondary reactions.
Peptide Purity: A peptide purity of 75% is generally sufficient for antibody generation and testing. However, 95% purity is recommended for studying biological activity.
Peptide Solubility: The amino acid composition of a peptide strongly influences its solubility. Ideally, the hydrophobic amino acid content should be kept below 50%, and there should be a minimum of one charged residue for every five amino acids.

Amino acid classifications
Hydrophobic (non-polar) amino acids: Ala, Ile, Leu, Met, Phe, Trp, Val
Uncharged (polar) amino acids: Asn, Cys, Gly, Gln, Pro, Ser, Thr, Tyr
Acidic (polar) amino acids: Asp, Glu
Basic (polar) amino acids: His, Lys, Arg
Peptides with a large number of hydrophobic amino acids have limited solubility in aqueous solutions; some are insoluble completely. These peptides are commonly difficult to use experimentally and can be difficult to purify. At physiological pH, acidic (Asp, Glu) and basic (Lys, Arg) amino acids all have charged side chains. Some hydrophobic amino acids located in nonessential positions could be substituted for conservative amino acids such as alanine or glycine, removed altogether, or replaced with analogs. In addition, a single conservative substitution or the addition of some polar residues to the N- or C-terminus of a peptide could improve its solubility.
There are a number of simple characteristics that can be used to predict solubility:
- Peptides <5 amino acids in length are commonly soluble in aqueous solutions. However, the whole sequence is only hydrophobic amino acids it will have limited solubility, or could even be completely insoluble.
- Hydrophilic peptides that contain >25% charged residues (Glu, Gln, Lys, Arg, and His) and hydrophobic peptides that consist of ≥50% hydrophobic amino acids might be insoluble or only partly soluble in aqueous solutions. Such peptides could be dissolved in organic solvents such as dimethylsulfoxide (DMSO) if they do not contain C, W, or M, or ethanol, dimethylformamide (DMF), acetonitrile, urea, isopropyl alcohol, acetic acid, or 4–8 M guanidine hydrochloride (GdnHCl). They can then be diluted carefully into aqueous solutions.
- Hydrophobic peptides that contain >75% hydrophobic residues do not commonly dissolve in aqueous solutions. Instead, very strong solvents, such as TFA or formic acid, must be used for the initial solubilization step. However, these peptides might precipitate when added to buffered aqueous solutions. If this occurs, high concentrations of organic solvent or denaturants might be required to dissolve these peptides.
- Peptides that include a very high percentage (>75%) of Glu, Asn, His, Lys, Gln, Arg, Ser, or Thr residues can form intermolecular hydrogen bonds (cross-links); therefore, they can form gels in concentrated aqueous solutions. Therefore, these peptides should be dissolved in organic solvents. It is important that the initial solvent selected is compatible with the final experiment. After the peptides have been dissolved in organic solvent, the solution should be added slowly (dropwise) to a stirred aqueous buffer solution. If the resulting solution begins to become turbid, the solubility limit has been reached.
Challenging Amino Acids
- Cysteine (C) and methionine (M)can be oxidized rapidly, which can inhibit the cleavage of the protecting groups during peptide synthesis and purification. Methionine can be oxidized irreversibly into methionine sulfoxide, which subsequently forms methionine sulfone. Cysteine is oxidized more rapidly at higher pH. Its thiol group is deprotonated easily, and forms intra- and inter-chain disulfide bonds readily. As such, sequences should be designed to contain as few C and M residues as possible, and conservative replacements can sometimes be made. For example, cysteine can be replaced with serine, and norleucine (Nle) can be used instead of methionine. When selecting a peptide from the native protein, separating two Cys residues into separate peptides allows improved synthesis and a final product that is more pure. This is because multiple cysteines on the same peptide are likely to form disulfide linkages. To prevent this, a reducing agent such as dithiothreitol (DTT), or tris(2-carboxyethylphosphine) hydrochloride (TCEP) should be added to the buffer. Alternatively, the cysteines should be replaced with serine residues. Cysteines can affect antibody activity. Because free cysteines are rare in vivo, they might not be recognized as part of the native peptide structure.
- Glutamine (Q) is unstable. Glutamine residues located at the N terminus of a peptide or protein can form cyclic pyroglutamate under the acidic conditions used to protect group cleavage. This can be prevented by acetylating an N-terminal glutamine, or by substituting glutamine with pre-formed pyroglutamic acid or a conservative amino acid.
- The asparagine (N) should be avoided. An N-terminal asparagine protecting group can be challenging to remove during cleavage. Therefore, it should be removed or replaced with another amino acid.
- Aspartic acid (D) is highly susceptible to dehydration and can form a cyclic imide intermediate. The cyclic imide intermediate can be hydrolyzed and cause peptide cleavage under acidic conditions when it is paired with glycine, serine, or proline. Ultimately, all aspartate within a given compound can be converted completely into iso-aspartate. These combinations should be avoided wherever possible by using substitutions or by modifying the sequence.
- Multiple serine (S) or proline (P) residues can cause significant deletions during peptide synthesis. This is particularly true with proline residues, which can undergo cis/trans isomerization and consequently reduce peptide purity.
- A sequence of glutamine, isoleucine, leucine, phenylalanine, threonine, tyrosine, or valine residues might result in beta-sheet formation. This could prevent a growing peptide from dissolving completely during synthesis, leading to the deletion of sequences. Therefore, multiples of the above amino acids in an adjacent pattern should be avoided if possible. If such sequences cannot be avoided, the beta-sheets structure can be prevented by conservative substitutions such as Asn for Gln or Ser for Thr, by the addition of Gly or Pro every third amino acid, or by moving the sequence.
- Glycine (G) as the third amino acid from the N-terminus, particularly if Pro or Gly is in position 1 or 2, can form diketopiperazine and pyroglutamic acid. This combination should be avoided whenever possible.
Considerations for Carrier Protein Coupling:
Although coupling strategies often vary according to the sequence of the peptide, the carrier proteins should be linked to the peptide via C- or N-terminal residues. If there are no internal cysteine residues, then a cysteine should be added to the sequence. Generally, we recommend that the carrier protein should be conjugated to the N-terminus of the peptide. Peptides of interest are conjugated to carrier proteins that contain many epitopes to stimulate T-helper cells because most peptides alone are too small to elicit an immune response large enough to lead to antibody generation. The T-helper cells induce the B-cell response that generates the antibodies. The immune system recognized the peptide-carrier protein complex as though it was a whole protein. Therefore, some antibodies target the linker region and the carrier protein itself. Nevertheless, these non-specific antibodies can be removed during purification. There are several common carrier proteins that are used during antibody production.
- KLH (keyhole limpet hemocyanin) is a copper-containing protein found in arthropods and mollusks. It is isolated from Megathura crenulata and has a MW of 4.5 x 10^5-1.3 x 10^7 Da. KLH is the most commonly used carrier because of its excellent immunogenicity in rabbits. The peptide-KLH is usually used for immunization, while peptide-BSA is used for ELISA testing.
- BSA (bovine serum albumin), a plasma protein in cattle, is one of the most stable and soluble albumins. It has a MW of 67 x 10^3 Da and it contains 59 lysines. About 30-35 of these primary amines can be used for linker conjugation. If the BSA is used as a blocking buffer reagent, antisera against peptide-BSA conjugates can generate false positives.
- OVA (ovalbumin) is a protein isolated from hen egg whites. Its MW is about 45 x 10^3 Da. It makes a good second carrier protein if antibodies are specific for the peptide.
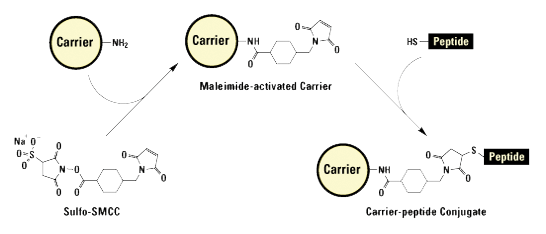
All modifications carrying thiol-reactive functional groups can be used. There are three commonly used reactive groups:
- Iodoacetamides
- Maleimides
- Alkyl halides
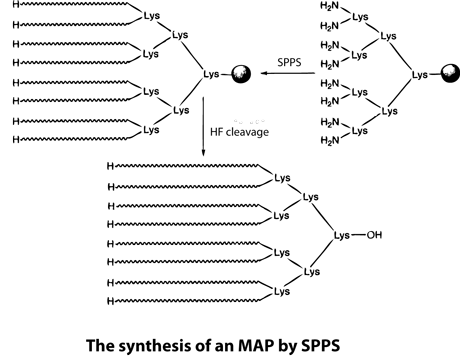
Multiple antigenic peptides (MAPs) are artificially branched peptides. Lysine residues are used as the scaffolding core to form up to eight branches that have the same or different peptide sequences. MAPs have been used for antibody production in immunological studies for a long time. Although some peptides render a lower immunological response, new structures, such as concentrated branched peptides, can increase the immunological response dramatically. Many antigenic peptides can be synthesized using standard SPPS. During this process, Boc-Lys(Boc)-OH is anchored to a resin, the peptides are treated with TFA, and cycles of deprotection and coupling are then performed sequentially. Peptides to be used in immunological studies are then synthesized as each of the eight branches. However, the nature of branched proteins is unknown, and so MAP synthesis can be challenging. For example, the space between the eight branches can cause the peptides to aggregate on the resin, which results in low coupling yields and/or deletions.
. To overcome this problem, LifeTein uses PeptideSyn technology for chemical ligation. This allows the desired peptide dendrimer to be formed at higher yields than could be achieved using traditional methods. The branched structures result in a higher molecular weight of the protein, which aids immunogenicicty.
Peptide Modifications: An important advantage of synthetic peptides is that they can be synthesized into exact conformations or according to the specific characteristics that are required for the end application. Peptides in general and certain amino acids contain distinct moieties that can be modified: an N-terminal amino group, a C-terminal carboxyl group, the alpha-amino group of lysines, the hydroxyl group of serine, threonine, and tyrosine residues, arginine residues containing guanidine groups, and the thiol group of cysteine.
A. N-terminal amino group
B. C-terminal carboxyl group
C. alpha-Amino group on lysine
D. Hydroxyl group on serine, threonine, and tyrosine
E. Guanidine group on arginine
F. Thiol group on cysteine

Although the list shown above includes the most common modifications, it is by no means complete. Some modifications occur post-translationally in vivo, whereas others take the form of the substitution of natural amino acids for non-natural variants. In addition, specific tags or proteins can be cross-linked to the moieties described above. Because of the C-to-N direction of peptide synthesis, we recommended that tags and dyes be conjugated to the N-terminus of the peptide so that only the full-length sequences are labeled.
LifeTein Antibody Guarantees
- ELISA results of 1:32,000 for any peptide polyclonal antibody project
- No cross-reactivity for phosphopeptide-specific polyclonal projects, and a guaranteed 1:32,000 ELISA titer
- ELISA titers of 1:80,000 for all peptide monoclonal antibody projects
- Two positive clones for allmonoclonal antibody projects
|
|
|
|
Reference:
Jia Shen. et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497, 383–387 (16 May 2013), doi:10.1038/nature12080
LifeTein helped designed and synthesized a series of phosphorylated peptides. Then the peptides were used for phospho-specific antibody productions. The phospo-specific antibodies by LifeTein were confirmed to react with the epidermal growth factor receptor (EGFR). The Hung's lab showed that AGO2-Y393 phosphorylation mediates EGFR-enhanced cell survival and invasiveness under hypoxia. These findings suggest that modulation of miRNA biogenesis is important for stress response in tumour cells.
... The following peptides were chemically synthesized for antibody production in mice (Lifetein Conc.), Elisa verification (LifeteinConc.) and peptide competition assay in immunohistochemistry (IHC)...
Supplementary information